MMB-BioIT
The Medical Microbiology bioinformatics (MMB-BioIT) team from the University Medical Center Utrecht (UMCU) consists of researchers with different scientifical backgrounds. Our efforts are dedicated to develop and use bioinformatic tools that facilitate data analysis on large amounts of DNA sequences, including whole genome sequences (WGS), taxonomic profiling using 16S rRNA gene sequencing and metagenomic sequencing.

Highlights

Population Genetics
The worldwide emergence of antibiotic resistant bacteria causing difficult to treat infections highlights the importance to track their local and global transmission. Using WGS or metagenomic sequences, different bioinformatic tools are used to group these bacteria based on the core and/or accessory genome, including mobile genetic elements encoding e.g. antibiotic resistance genes. In addition, we developed tools to reconstruct plasmids based on short read sequences in order to determine transmission patterns of mobile genetic elements.
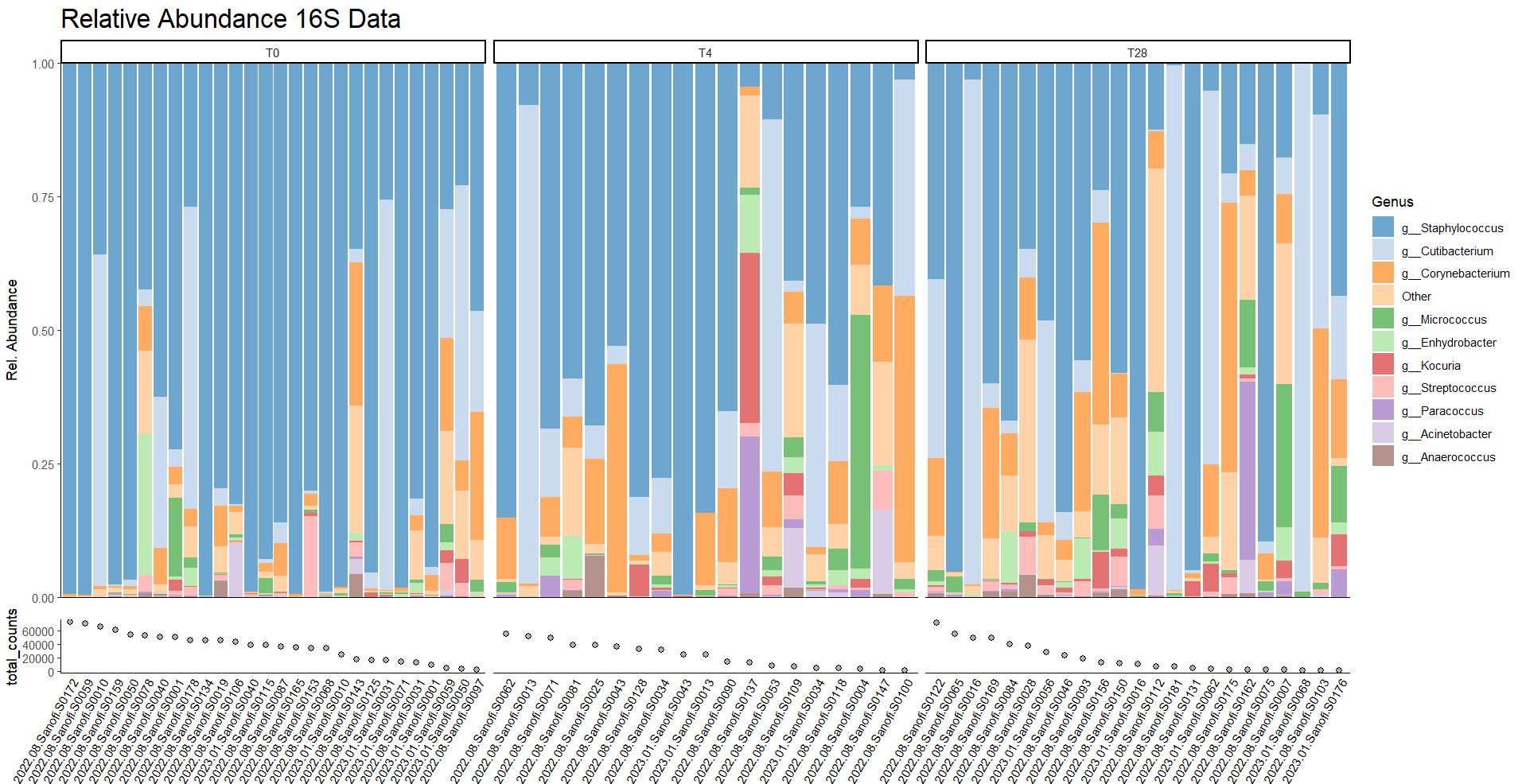
Microbiome analysis
A microbiome is a collective polymicrobial community, or ‘microbiota’, including bacteria, archaea, algae, protozoa, fungi, and viruses. Over the last decades research has shown that microbiome composition can be linked to health and disease. In our group we perform microbiome analysis on taxonomic level using 16S rRNA or metagenomic sequencing. The latter can also be used to analyze metabolic pathways encoded by bacteria.
Our Tools
The mission of our team is that the developed tools and analyzed data are released to enable reproducible computational biology analyses following the FAIR (Findable, Accessible, Interoperable and Reusable) principles. Find more about our tools in the link below.

