Tools & Education
In MMBioIT we actively develop tools for bacterial genomics, focusing on providing software that is easy to install and use. We strongly believe in transparent and collaborative science, therefore all our tools are completely open-source. We encourage you to try our different tools.
Tools
Whole Genome Sequencing
 gplascc
gplasccgplas is a tool to bin plasmid-predicted contigs based on sequence composition, coverage and assembly graph information. This tool extends the possibility of accurately binning predicted plasmid contigs into several discrete plasmid components.
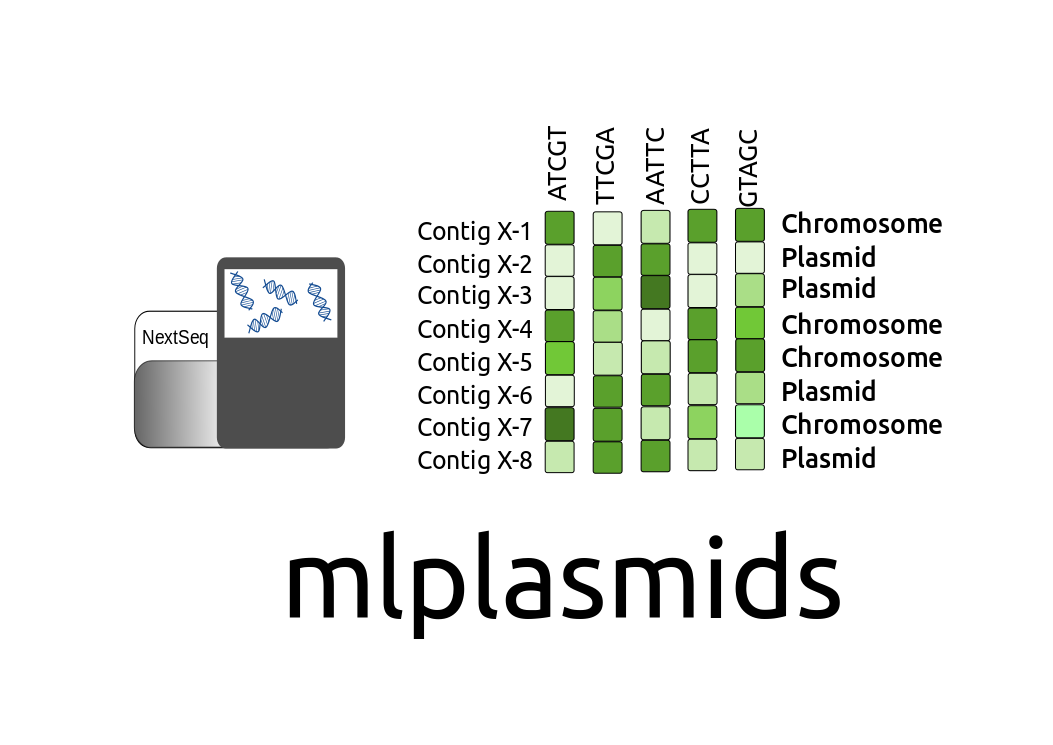 mlplasmids
mlplasmidsmlplasmids consists of binary classifiers to predict contigs either as plasmid-derived or chromosome-derived. It currently classifies short-read contigs as chromosomes and plasmids for Enterococcus faecium, Enterococcus faecalis, Klebsiella pneumoniae and Acinetobacter baumannii.
 plasmidEC
plasmidECplasmidEC identifies plasmid-derived contigs in all bacterial species. This tool combines the output of three binary classification tools and implements a majority vote system. Currently, the user can choose any combination of the following four softwares: Centrifuge, Platon, RFPlasmid and mlplasmids.
 plasmidCC
plasmidCCplasmidCC is a plasmid classification tool that uses Centrifuge to predict the origin of contigs (plasmid or chromosome). Out of the box, plasmidCC can be used to predict plasmid contigs of a selected number of species. By supplying a custom Centrifuge database, plasmidCC can be used for any desired species.
 Bactofidia
Bactofidiabactofidia is a bacterial assembly and basic analysis pipeline using Snakemake and bioconda. It is currently written for paired-end Illumina data with length 250 or 150. The pipeline is written to ensure reproducibility, and creates virtual software environments with the software versions that are used for analysis.
 ClonalTraker
ClonalTrakerClonalTraker is a tool to compare any two vancomycin-resistant Enterococcus faecium (VRE), and the genomic context of the van operon.
Metagenomics
 MetaMobilePicker
MetaMobilePickerMetaMobilePicker is a computational pipeline designed to identify mobile genetic elements and antimicrobial resistance genes in metagenomics samples. Currently it identifies plasmids, insertion sequences and bacteriophages.
Tourmaline is an amplicon sequence processing workflow for Illumina sequence data developed by the Nacional Oceanic and Atmospheric Administration that uses QIIME 2 and the software packages it wraps. We modified this package to fit our sequencing data and system requirements.
Educational resources
 Molecular Epidemiology of Infectious Diseases - computer practical
Molecular Epidemiology of Infectious Diseases - computer practicalThree day graduate level course on molecular epidemiology teaching the command line, WGS, annotation and pangenomics.
 Microbial Genomics Workshop
Microbial Genomics WorkshopThree day course on microbial genomics from commandline to SNP analyses using data from the Lenski Long Term Evolution experiment.
